Sequencing and Annotating the Wild Rice Genome
We have been diligently working to develop the first annotated, de novo assembly of the wild rice genome. We have used PacBio and Illumina technologies to generate sequence data as well as HiC and Chicago libraries to aid in the assembly of the genome. Annotation of the genome is currently underway using the transcriptome analysis of six different wild rice tissue types including root, shoot, leaf, seed, female florets, and male florets.
RNA Sequencing
RNA-Seq can be used to define when and how strongly genomic loci are expressed within a cell over time. The current aim for RNA-Seq in the Kimball program is to assess gene expression in NWR as it relates to dormancy. RNA extracted from seeds over a 9-month period, starting post-harvest, will be run through an RNA-Seq workflow to determine when different genomic loci are expressed throughout the dormancy cycle.
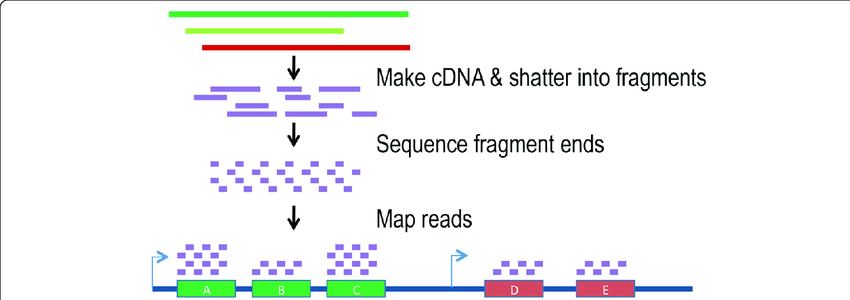
Genotyping by Sequencing and SNP Marker Identification
The Kimball program is working to optimize genotyping-by-sequencing (GBS) for use in Northern Wild Rice research. GBS is a type of DNA sequencing that reduces genome complexity to capture and sequence portions of the genome that are most interesting to plant scientists. GBS accomplishes this using restriction enzymes to create smaller fragments of DNA. Restriction enzymes make cuts in strands of DNA at sequences known as genetic palindromes, or sequences that read the same on both the forward and reverse strands. Adapters are then ligated, or joined, onto the “sticky” cut ends of digested fragments and then the fragments are amplified by polymerase chain reaction (PCR), making many copies of the DNA sequence, prior to sequencing.
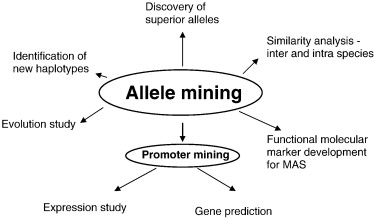
Allele Mining
Allele Mining refers to the process of identifying and using genetic variation that affects a plant's phenotype (the physical manifestation of a trait, such as flower color). NWR is a diploid, which means it has two different copies of each gene. Each of these copies is called an allele and these alleles can have different effects on the plant's phenotype. In cultivated NWR, reduced grain shattering, higher yield, and disease resistance are some examples of desirable traits. Genes controlling these traits are typically identified by their association with genetic markers, such as single nucleotide polymorphisms (SNPs), which are identified using technology such as genotyping-by-sequencing (GBS).
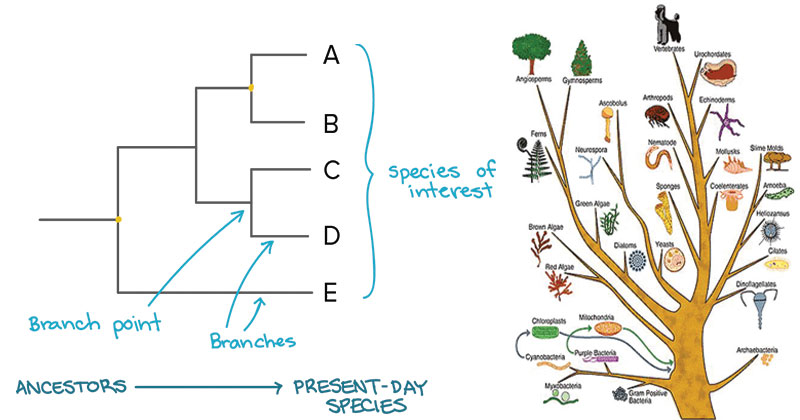
Comparative Genomics
All life on earth is descended from a common ancestor. Major branches on the tree of life, such as those at the point where plant and animal lineages diverged from each other, occurred in the distant past. The divergence of different plant genera happened in the much more recent past relative to the age of the earth. For example, NWR (Zizania palustris) and white rice (Oryza sativa) are both considered to be “rice” in the broad sense as they are both members of the tribe Oryzeae, but the two genera, Zizania and Oryza last shared a common ancestor approximately 26.7 million years ago. Since the two genera diverged relatively recently from an evolutionary point of view, we expect the two genera to have very similar genomes. On the other hand, 26.7 million years have elapsed since they last shared a common ancestor so there has been sufficient time for differences to accrue.