Dr. Kimball's Google Scholar Profile
Preprints
Mickelson, A., McGilp, L., and Kimball, J. (2023) Investigating Experimental Storage Methodologies for the Understudied Intermediate Recalcitrant Seed of Northern Wild Rice (Zizania palustris L.). bioRxiv 10.1101/2023.08.03.551837 https://www.biorxiv.org/content/10.1101/2023.08.03.551837v1
Haas, M., McGilp, L., Shao, M., Millas, R., Castel-Miller, C., Shanon, LM., Kern, A., and Kimball, J (2022). Genetic Analysis of Wild and Cultivated Populations of Northern Wild rice (Zizania palustris L.) Reveal New Insights into Gene Flow and Domestication. bioRxiv 2022.08.25.505308 https://www.biorxiv.org/content/10.1101/2022.08.25.505308v1.full
Gietzel, C., McGilp, L., and Kimball, J. (2022) Spatiotemporal Profiling of Seed-Associated Microbes of an Aquatic, Intermediate Recalcitrant Species, Zizania palustris L. and the Impact of Anti-Microbial Seed Treatments. bioRxiv 2022.04.20488936 http:/www.biorxiv.org/content/10.1101/2022.04.20.488936v1
2023
McGilp, L., Castell-Miller, C., Haas, M., Millas, R., & Kimball, J. (2023). Northern Wild Rice (Zizania palustris L.) breeding, genetics, and conservation. Crop Science, 00, 1– 30. https://doi.org/10.1002/csc2.20973
2022
McGilp, L., Semington, A., and Kimball, J. (2021) Dormancy Breaking Treatments in Northern Wild Rice (Zizania palustris L.) Seed Suggest a Physiological Source of Dormancy. Journal of Plant Growth Regulation. https://doi.org/10.1007/s10725-022-00859-0
Eizenga G.C., Kim H., Jung J.K.H., Greenberg A.J., Edwards J.D., Naredo M.E.B., Banaticla-Hilario M.C.N., Harrington S.E., Shi Y., Kimball J.A., Harper L.A., McNally K.L., McCouch S.R. (2022) Phenotypic Variation and the Impact of Admixture in the Oryza rufipogon Species Complex (ORSC). Frontiers in Plant Science. https://doi.org/10.3389/fpls.2022.787703
2021
Gietzel, C., Duquette, J., McGilp, L., & Kimball, J. (2021). Evaluation of a Recessive Male Floret Color in Cultivated Northern Wild Rice (Zizania palustris L.) for Pollen-Mediated Gene Flow Studies. Crop Science, 1-11. https://doi.org/10.1002/csc2.20647
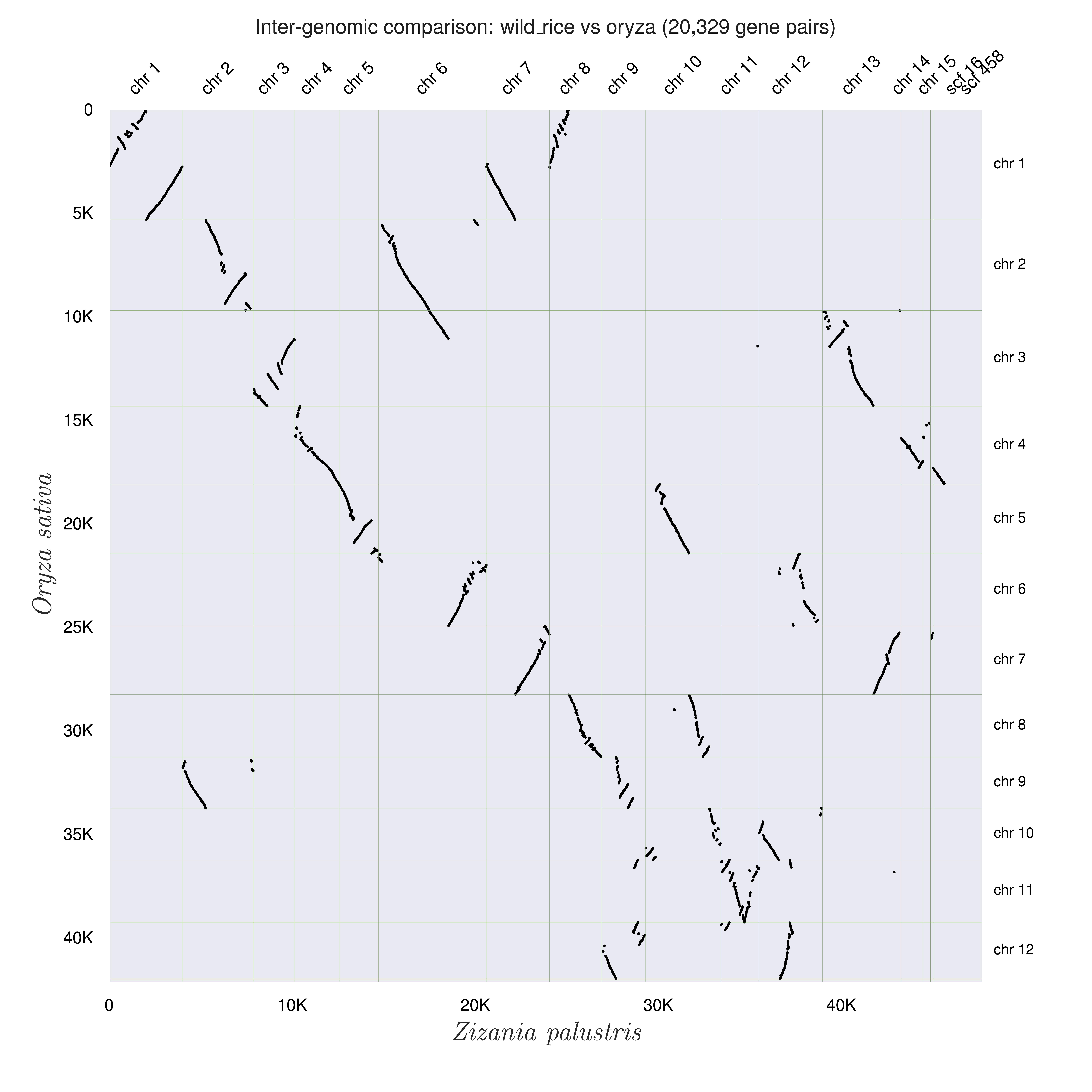
Haas, M., Kono, T., Macchietto, M., Millas, R., McGilp, L., Shao, M., Duquette, J., Hirsch, C.N., and Kimball, J. (2021) Whole Genome Assembly and Annotation of Northern Wild Rice, Zizania palustris L., Supports a Whole Genome Duplication in the Zizania Genus. Plant J, 107: 1802-1818. https://doi.org/10.1111/tpj.15419
2020
McGilp, L., Duquette, J., Braaten, D., Kimball, J., & Porter, R. (2020). Investigation of Variable Storage Conditions for Cultivated Northern Wild Rice and their Effects on Seed Viability and Dormancy. Seed Science Research, 30(1), 21-28. https://doi:10.1017/S0960258520000033
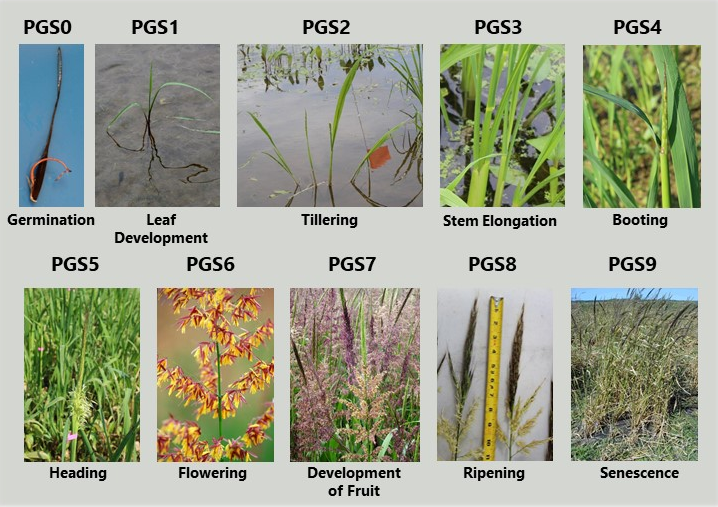
Duquette, J. and Kimball, J.A. (2020). Phenological Stages of Cultivated Northern Wild Rice According to the BBCH Scale. Ann Appl Biol. 2020; 176:350-356. https://doi.org/10.1111/aab.12588
Tuleski, T. R., Kimball, J., do Amaral, F. P., Pereira, T. P., Tadra-Sfeir, M. Z., de Oliveira Pedrosa, F., ... & Stacey, G. (2020). Herbaspirillum rubrisubalbicans as a Phytopathogenic Model to Study the Immune System of Sorghum bicolor. Molecular Plant-Microbe Interactions, 33(2), 235-246. https://doi.org/10.1094/MPMI-06-19-0154-R
2019
Kimball, J.A., Y. Cui, D. Chen, P. Brown, W. Rooney, G. Stacey, and P. Balint-Kurti. 2019. Identification of QTL for Target Leaf Spot Resistance in Sorghum bicolor and Investigation of Relationships Between Disease Resistance and Variation in the MAMP Response. Scientific Reports 9: 18285 doi:10.1038/s41598-019-54802-x
Samira, R., X. Zhang, J. Kimball, Y. Cui, G. Stacey, and P.J. Balint-Kurti. 2019. Quantifying MAMP-Induced Production of Reactive Oxygen Species in Sorghum and Maize. Bio-101. doi.org/10.21769/BioProtoc.3304
Shao, M., M. Haas, A. Kern, and J.A. Kimball. 2019. Identification of Single Nucleotide Polymorphism Markers for Population Genetic Studies in Zizania palustris L. Conservation Genetic Resources, 12(3), 451-455. doi.org/10.1007/s12686-019-01116-9
2018
Yu, X. Kimball, J.A. and Milla-Lewis, S.R. 2018. High density genetic maps of St. Augustinegrass and Applications to Comparative Genomic Analysis and QTL Mapping for Turf Quality Traits. BMC Plant Biology. 18: 346.
Kimball, J.A., T.D. Tuong, E. Carbajal, and S.R. Milla-Lewis. 2018. Identification of QTL for cold tolerance-related traits in St. Augustinegrass. Molecular Breeding 38:67.
Kimball, J.A., T.D. Tuong, D.P. Livingston, and S.R. Milla-Lewis. 2018. Assessing freeze-tolerance in St. Augustinegrass: acclimation effects. Euphytica 213:282.
2017
Kimball, J.A., T.D. Tuong, D.P. Livingston, and S.R. Milla-Lewis. 2017. Assessing freeze-tolerance in St. Augustinegrass: temperature response and evaluation methods. Euphytica 213:110. DOI 10.1007/x10681-017-1899-z
2016
Kim, H., J. Jung, N. Singh, A. Greenberg, J.J. Doyle, W. Tyagi, J.A.Kimball, R. Sackville-Hamilton, and S.R. McCouch. 2016. Population dynamics among six major groups of the Oryza rufipogon complex, wild relative of cultivated Asian rice. Rice 9:56.
Kimball, J.A., M.C. Zuleta, E. Carbajal, and S.R. Milla-Lewis. 2016. Combining ability of cold tolerance and turf quality-related traits in St. Augustinegrass. Hort Sci 51:810-815.
2013
Kimball J.A., M.C. Zuleta, K.E. Kenworthy, and S.R. Milla-Lewis. 2013. Use of molecular markers for identifying off-types and contaminants in samples of St. Augustinegrass cultivar ‘Captiva’. International Turfgrass Society Research Journal, Vol. 12:267-274.
Milla-Lewis, S.R., J.A. Kimball, T.D. Tuong, T.E. Claure, and D.P. Livingston. 2013. Freezing tolerance and histology of recovering nodes in St. Augustinegrass. International Turfgrass Society Research Journal, Vol. 12:523-530.
Imai, I., J.A. Kimball, B. Conway, K.M. Yeater, S.R. McCouch, and A. McClung. 2013. Validation of yield enhancing QTL from a low-yielding wild ancestor of rice. Mol. Breeding 32:101-120.
Kimball J.A., M.C. Zuleta, K.R. Harris-Schultz, K.E. Kenworthy, V.G. Lehman, and S.R. Milla-Lewis. 2013. Genetic relationships in Zoysia and the identification of putative interspecific hybrids using simple sequence repeat markers and inflorescence traits. Crop Sci. 53(1): 285-295.
2012
Kimball J.A., M.C. Zuleta, and S.R. Milla-Lewis. 2012. Assessment of molecular variability within the St. Augustinegrass cultivar ‘Raleigh’ using amplified fragment length polymorphism markers. HortSci. 47:1-6.
Kimball J.A., M.C. Zuleta, K.E. Kenworthy, V.G. Lehman, and S.R. Milla-Lewis. 2012. Assessment of Genetic Diversity in Zoysia Species using Amplified Fragment Length Polymorphism Markers. Crop Sci. 52(1):383-392.
2011
Milla-Lewis S.R., K.R. Harris-Schultz, M.C. Zuleta, J.A. Kimball, B.M. Schwartz, and W.W. Hanna. 2011 Use of sequence-related amplified polymorphism (SRAP) markers for comparing levels of genetic diversity in centipedegrass (Eremochloa ophiuroides (Munro) Hack.) germplasm. Genet. Resour. Crop Ev. DOI 10/1007/x10722-011-9780-8.
Ali M.L., A. McClung, M. Jia M, J. Kimball, S. McCouch, and G. Eizenga. 2011. A Rice Diversity Panel evaluated for genetic and agro-morphological diversity between subpopulations and its geographic distribution. Crop Sci. 51:2021-2035.
2010
Tung C.W., K. Zhao, M.H. Wright, M.L Ali, J. Jung, J. Kimball, W. Tyagi, M.J. Thomson, K. McNally, H. Leung, H. Kim, S.N. Ahn, A. Reynolds, B. Scheffler, G. Eizenga, A. McClung, C. Bustamante, and S.R. McCouch. 2010. Development of a research platform for dissecting phenotype-genotype associations in rice (Oryza spp.). Rice 3:205-217.
Zhao K., M. Wright, J. Kimball, G. Eizenga, A. McClung, M. Kovach, W. Tyagi, M.L. Ali, C.W. Tung, A. Reynolds, C.D. Bustamante, and S.R. McCouch. 2010. Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLOS One 5(5): e10780. doi:10.1371/journal.pone.0010780.
Orjuela J., A. Garavito, M. Bouniol, J.D. Arbelaez, L. Moreno, J. Kimball, G. Wilson, J.F. Rami, J. Tohme, S.R. McCouch and M. Lorieux. 2010. A universal core genetic map for rice. Theor. Appl. Genet. 120(3): 563-572.